Welcome to our website
A major roadblock in early drug design is the need to discover novel and potent small-molecule drug leads. Many projects do not achieve even one sub-micromolar lead and, in those that do, a single lead is typically obtained with nanomolar potency after several years of expert-led iterative optimisation work (thus, requiring extensive human and financial resources). Therapeutic targets are molecular, e.g. a disease-causing protein, or phenotypic, e.g. a cancer cell line or bacterial culture. For any of these targets, the more drug leads are discovered, the greater the likelihood that at least one will not have any preclinical issue and thus will eventually reach clinical trials.
Drugs overcoming clinical trials still face additional challenges after approval. In cancer, tumour heterogeneity and other inter-patient differences result in large response variability across patients despite having the same cancer type and being administered the same drug treatment. Precision oncology tackles this problem by attempting to identify drug-resistant patients early, so that they can be administered more effective therapies and they do not suffer unnecessary side effects. Unfortunately, most cancer drugs do not have response markers and most existing markers are far from being sufficiently predictive.
Our research aims at developing and applying computational methods to boost biomedical discoveries in these areas. We leverage three timely opportunities: artificial intelligence (AI) innovation, big data availability, and mature domain knowledge. This website introduces ourselves and the research we carry out in these fascinating areas.
Methods
In 2013, we showed how molecule-cell line datasets can be modelled with machine learning. We also demonstrated that machine learning can predict drug synergy on cancer cell lines as long as data inconsistencies between screening centres are taken into account. We also found evidence suggesting that estimating the uncertainty of individual predictions will be an effective way to enhance virtual screening on cancer cell lines. Ongoing research also reveals that graph neural networks outperform other algorithms.
* Menden, M. P.; Iorio, F.; Garnett, M.; McDermott, U.; Benes, C. H.; Ballester, P. J.; Saez-Rodriguez, J. PLoS One 2013, 8 (4), e61318.* Sidorov, P.; Naulaerts, S.; Ariey-Bonnet, J.; Pasquier, E.; Ballester, P. J. Frontiers in Chemistry 2019, 7, 509.
* Hernández-Hernández, S.; Vishwakarma, S.; Ballester, P. J. Proceedings of Machine Learning Research 2022, 179, 1–17.
* Ballester, P. J. Nature 2023, 624, 252.
Discovering novel antibiotics for human pathogens is a priority to face the challenge posed by the growing antimicrobial resistance trend. We thus investigate chemistry-informed AI models to screen ultralarge libraries for novel antibiotics against major human pathogens.
This is important to reveal the targets of phenotypic hits or better understand the polypharmacology and mechanism of action of drugs with already some known targets. We proposed a benchmark specific to target prediction and explain why the widespread use of virtual screening benchmarks is inadequate. These issues arise from not having the same objective: target prediction aims at finding the targets bound by a molecule, while virtual screening aims at finding the molecules binding to a target. We have also shown that reliability prediction boosts performance and provided a webserver implementing the method, which we are currently updating.
* Peón, A.; Dang, C. C.; Ballester, P. J. Frontiers in Chemistry 2016, 4, 15.* Peón, A.; Naulaerts, S.; Ballester, P. J. Scientific Reports 2017, 7 (1), 3820.
* Peón, A.; Li, H.; Ghislat, G.; Leung, K. S.; Wong, M. H.; Lu, G.; Ballester, P. J. Chemical Biology & Drug Design 2019, 94 (1), 1390–1401.
We developed USR, an ultrafast method to identify molecules with similar 3D shape to a template with activity for the target. We later provided a webserver to carry out virtual screening using this and related methods. We also made a hardware implementation of USR, which is even faster and more energy-efficient than these software implementations at the cost of being less versatile.
* Ballester, P. J.; Richards, W. G. Journal of Computational Chemistry 2007, 28 (10), 1711–1723.* Li, H.; Leung, K.-S.; Wong, M.-H.; Ballester, P. J. Nucleic Acids Research 2016, 44 (W1), W436–W441.
* Morro, A.; Canals V.; Oliver A.; Alomar, M.L.; Galán-Prado, F.; Ballester, P.J.; Rossello, J.L. IEEE transactions on neural networks and learning systems 29 (4), 1371-1375.
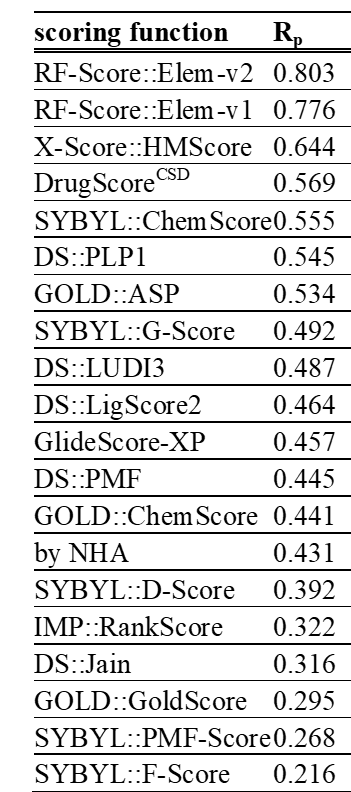
Also known as structure-based virtual screening, it is an effective way to discover potential drug leads from their docked poses against the 3D structure of the target. This is typically posed as a binary classification problem and the resulting models are called scoring functions. Virtual screening is hard due to extreme class imbalance and data biases. We showed how machine learning and data augmentation can improve scoring functions for virtual screening. Strong progress in machine learning for structure-based virtual screening has been made in recent years, also in the prospective applications of these methods. Scoring functions built with machine learning, instead of the classical ones assuming linearity between features and binding strength, provide higher hit rates while identifying molecules with higher potency too.
* Wójcikowski, M.; Ballester, P. J.; Siedlecki, P. Scientific Reports 2017, 7, 46710.* Li, H.; Sze, K.-H.; Lu, G.; Ballester, P. J. WIREs Computational Molecular Science 2021, e1478.
* Ghislat, G.; Rahman, T.; Ballester, P. J. Current Opinion in Chemical Biology 2021, 28–34.
* Ain, Q. U.; Aleksandrova, A.; Roessler, F. D.; Ballester, P. J. WIREs Computational Molecular Science 2015, 5 (6), 405–424.
* Fresnais, L.; Ballester, P. J. Briefings in Bioinformatics 2020, bbaa095.
We still cannot reliably estimate the prospective performance of a method from retrospective analysis. We have contributed to this effort by explaining how to reduce decoy bias or providing a user-friendly protocol to build and benchmark machine-learning scoring functions (In Press). Rigorous in-depth analyses are also important to make progress on this key problem.
* Ballester, P. J. Drug Discovery Today: Technologies. 2020, 81–87.* Tran-Nguyen, V.-K.; Ballester, P. J. Journal of Chemical Inf. and Model. 2023, 27-28.
* Tran-Nguyen, V.-K.; Junaid, M.; Simeon, S.; Ballester, P. J. Nature Protocols 2023 (In Press; doi:10.1038/s41596-023-00885-w).
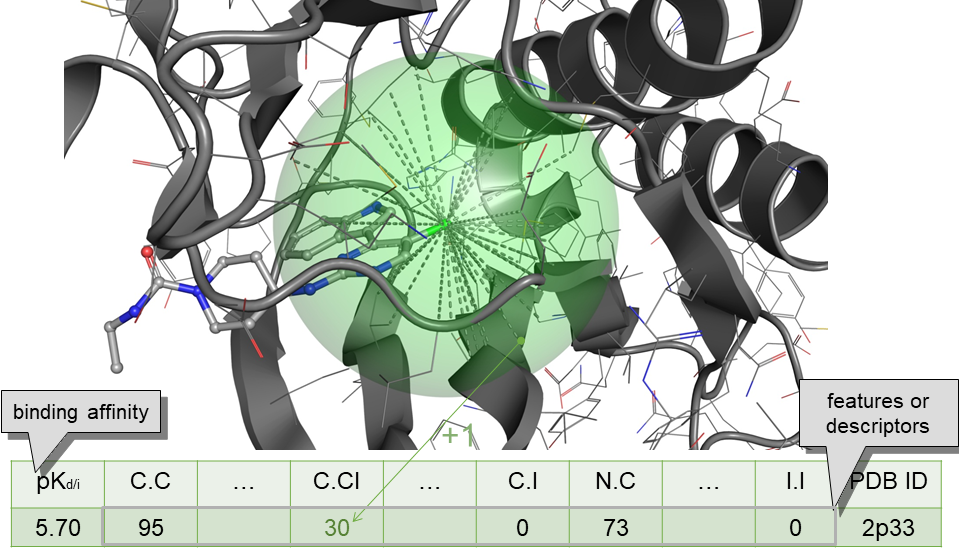
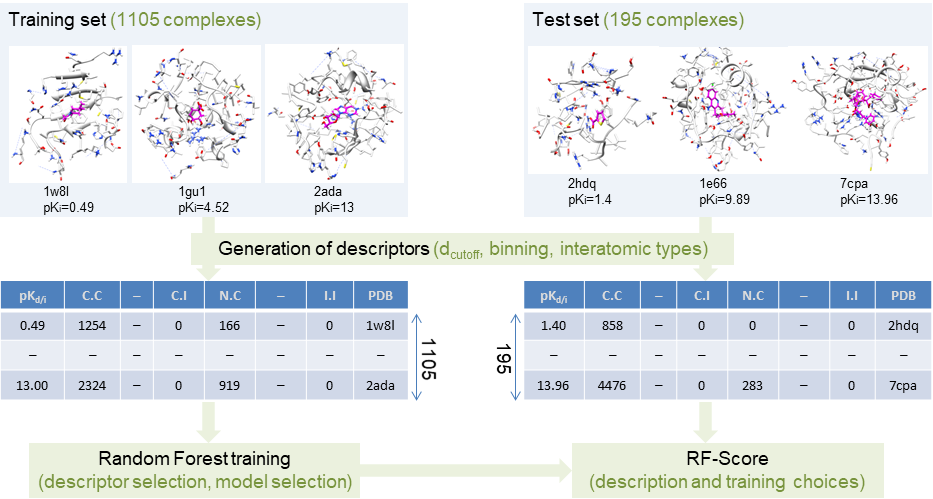
Not optimal for virtual screening, as non-binders are practically not considered. However, these structure-based models can be useful for optimising the potency of known small-molecule binders against a target. Having the least confounding factors, this type of data leads to the most robust results. In 2010, we discovered that machine-learning scoring functions are substantially more predictive than classical scoring functions at this task. This performance gap grows with larger training sets, which is still true when one removes test complexes with similar molecules or targets to those in the training set. We also found that expert-based selection of features, the innovation engine of classical scoring functions, is actually suboptimal for machine-learning scoring functions.
* Li, H.; Sze, K.-H.; Lu, G.; Ballester, P. J. WIREs Computational Molecular Science 2020, 10 (5), e1465.* Ballester, P. J.; Mitchell, J. B. O. Bioinformatics 2010, 26 (9), 1169–1175.
* Li, H.; Peng, J.; Sidorov, P.; Leung, Y.; Leung, K. S.; Wong, M. H.; Lu, G.; Ballester, P. J. Bioinformatics 2019, 35 (20), 3989–3995.
* Ballester, P. J.; Schreyer, A.; Blundell, T. L. Journal of Chemical Information and Modeling 2014, 54 (3), 944–955.
Machine-learning scoring functions as a part of holistic efforts to find other uses for existing drugs, for instance as anti-aging treatments.
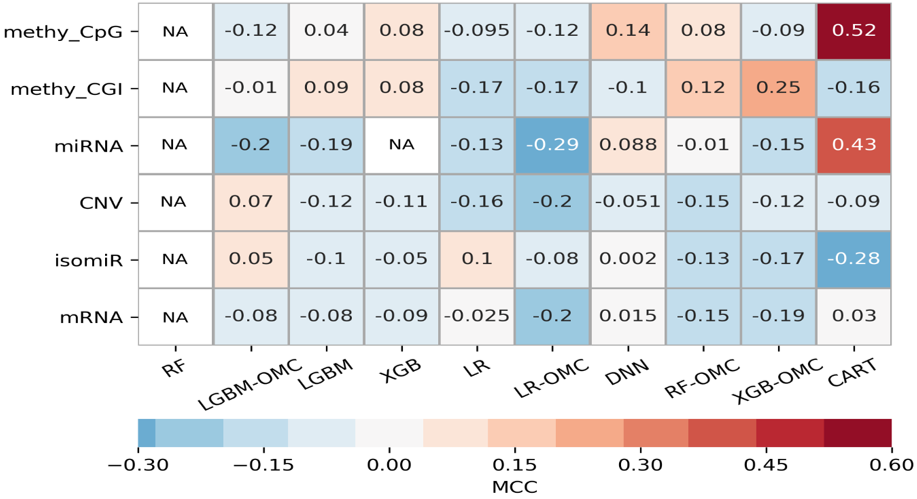
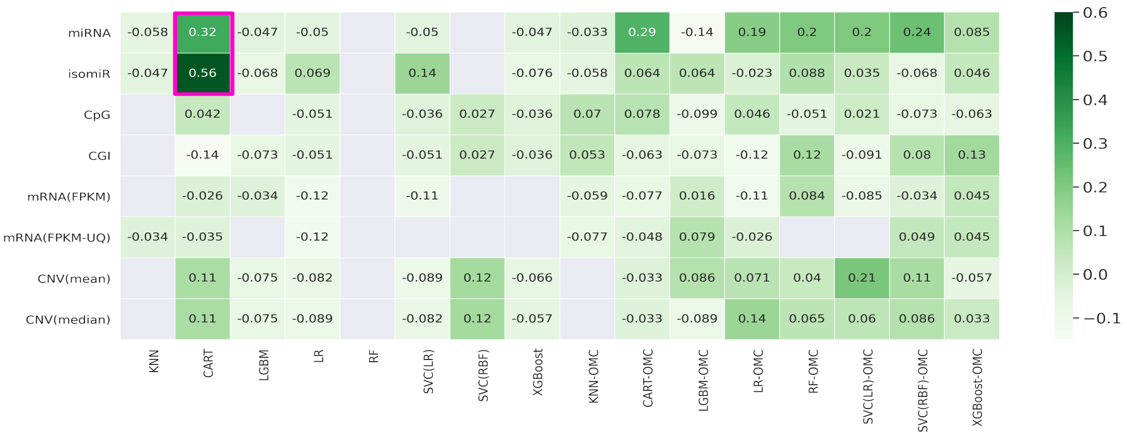
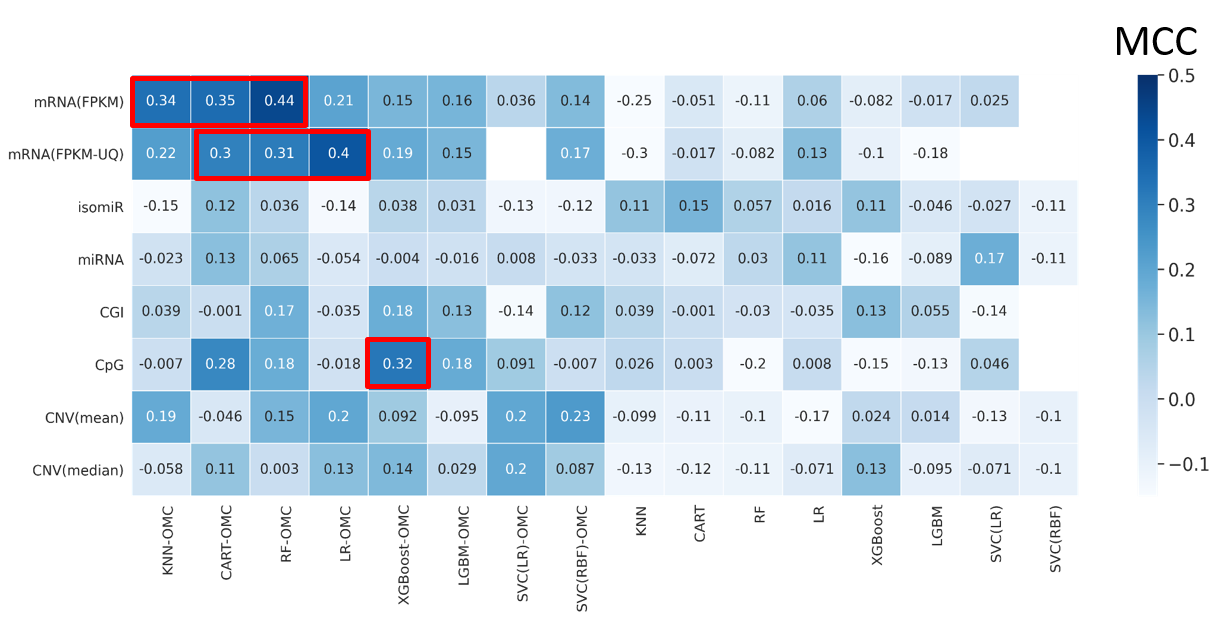
* Ziehm, M.; Kaur, S.; Ivanov, D. K.; Ballester, P. J.; Marcus, D.; Partridge, L.; Thornton, J. M. Aging Cell 2017.Cancer drugs are only effective in a fraction of patients. We thus need to predict which patients will be resistant to a given drug. Such prediction is based on the presence of a somatic mutation (mutation marker) or the a priori combined expression of expert-selected genes (gene expression signature), but considering other genes, profiles and model-building approaches improves the prediction. Especially in terms of recall. A promising alternative is using AI to predict how patients respond to a drug from the multiple molecular profiles (multi-omics) of their tumours. This AI approach has so far provided enhanced prediction in paclitaxel-treated breast cancer patients, doxorubicin-treated breast cancer patients, gemcitabine-treated pancreatic cancer patients and temolozomide-treated low-grade-glioma patients. Overall, we found that the best learning algorithm - molecular profile combination changes across drugs and cancer types and is also easy to miss as almost all other combinations are not predictive. This highlights the critical importance of considering many algorithms and profiles when analysing these clinical pharmaco-omic datasets.
* Ballester, P. J.; Stevens, R.; Haibe-Kains, B.; Huang, R. S.; Aittokallio, T. Briefings in Bioinformatics 2021, bbab450.* Naulaerts, S.; Dang, C. C.; Ballester, P. J. Oncotarget 2017, 8 (57), 97025–97040.
* Ballester, P. J.; Carmona, J. npj Precision Oncology 2021, 5 (1), 1–3.
* Bomane, A.; Gonçalves, A.; Ballester, P. J. Frontiers in Genetics 2019, 10, 1041.
* Ogunleye, A. Z.; Piyawajanusorn, C.; Gonçalves, A.; Ghislat, G.; Ballester, P. J. Advanced Science 2022, 9 (24), e2201501.
* Ogunleye, A. Z.; Piyawajanusorn, C.; Ghislat, G.; Ballester, P. J. Health Data Science (In Press).
Applications
Prospectively applying our developed methods is important as a reality check, understanding how to improve methods further and actually making biomedical discoveries. However, this requires finding a collaborator to carry out in vitro tests and also provide expertise on the target. We are hence keen to enter this type of collaborations and have been able to complete some over the years. These are some examples below (we do not include the prospective applications by others using our methods):
Screening over a billion make-on-demand compounds with our AI models has led to the identification of nanomolar inhibitors of colorectal and glioma cell lines (all with novel chemical scaffolds). In vitro tests are not yet completed.
The targets of these phenotypic hits will be determined with our target prediction method, which already discovered low-nanomolar targets of drugs such as mebendazole and actarit.
* Ariey‐Bonnet, J.; Carrasco, K.; Le Grand, M.; Hoffer, L.; Betzi, S.; Feracci, M.; Tsvetkov, P.; Devred, F.; Collette, Y.; Morelli, X.; Ballester, P.J.; Pasquier, E. Molecular Oncology 2020, 14 (12), 3083–3099.* Ghislat, G.; Rahman, T.; Ballester, P. J. Biomolecules 2020, 10 (11), 1–11.
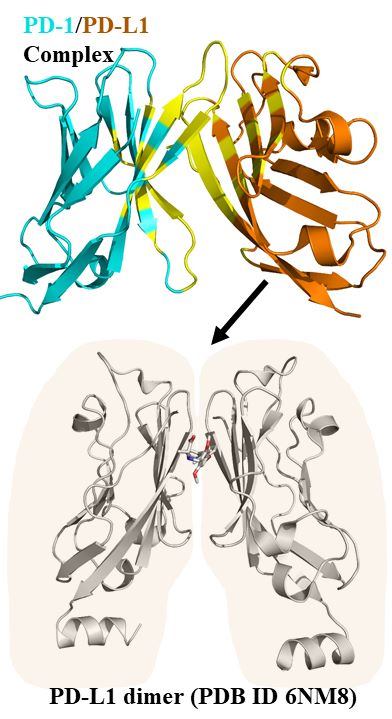
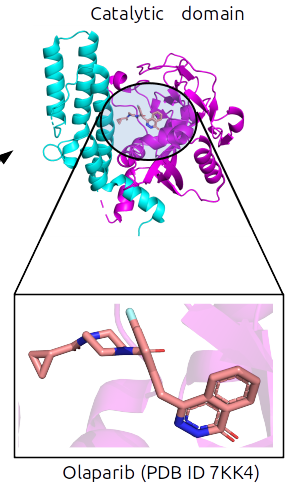
USR-based virtual screening has discovered novel low-micromolar inhibitors for a breast cancer target , a colorectal metastasis target and antibacterial targets, among others. This ligand-based approach was also successful when combined with a generic machine-learning scoring function on these antibacterial targets. We have also applied prospectively PARP1-specific machine-learning scoring functions, which have led so far to cell-active PARP1 small-molecule inhibitors as potent as 1 nanomolar.
* Ballester, P. J.; Westwood, I.; Laurieri, N.; Sim, E.; Richards, W. G. Journal of the Royal Society Interface 2010, 7 (43), 335–342.* Hoeger, B.; Diether, M.; Ballester, P. J.; Köhn, M. European Journal of Medicinal Chemistry 2014, 88, 89–100.
* Ballester, P. J.; Mangold, M.; Howard, N. I.; Robinson, R. L. M.; Abell, C.; Blumberger, J.; Mitchell, J. B. O. Journal of the Royal Society Interface 2012, 9 (77), 3196–3207.
On the other hand, we are keen on collaborating with clinicians in drug-based omics-informed precision oncology. The main obstacle to making an impact in this area is now clinical data sharing. Full data from two cohorts of patients with the same cancer type, with their tumours profiled with the same omics technology and treated with the same drug is hard to access. Collaborations providing such data to get our predictive models closer to the clinic is our priority. We would be also happy to consider other drug-cancer type cases for analysis if non-public data is available. Lastly, our AI protocols can be adapted to the analysis of longitudinal molecularly-profiled liquid-biopsy data for early detection of cancer. We are also keen to consider collaborators in this area.
Following a first degree in Astrophysics and an MSc in Information Processing and Neural Networks, I completed a PhD at Imperial College on geophysical data mining and inference. After that, I was awarded a Junior Research Fellowship at the University of Oxford and then held a short postdoc at the University of Cambridge, both associated with their Chemistry departments. In 2010, I was awarded a 4-year MRC Methodology Research Fellowship, which funded my staff scientist position at the European Bioinformatics Institute, and also a Governing Body Fellowship at Wolfson College Cambridge. In 2014, I moved to the south of France for a position as a tenured group leader at INSERM with A*MIDEX Excellence Chair and ANR Tremplin-ERC awards. In 2022, I returned to the UK as an Associate Professor at Imperial College London and Wolfson Fellow of the Royal Society.
Awards
• 2022-27 Royal Society Wolfson Fellow, UK.
• 2019-22 INSERM scientific excellence merit award (PEDR), France.
• 2017-19 ANR Tremplin - ERC, France.
• 2015-17 A*MIDEX Excellence Chair, France.
• 2014 Civil Servant Qualifying Exams (INSERM CR1), France.
• 2011-14 JRF & Governing Body Fellow, Wolfson College Cambridge, UK.
• 2010-14 MRC Methodology Research Fellow, UK.
• 2007-08 Junior Research Fellow (JRF), St Cross College Oxford, UK.
• 2000-01 Scholar, Sa Nostra Foundation, Spain.
• 2000 Civil Servant Qualifying Exams (Physics Innovation Expert), Spain.
Funding
I have been awarded a total of €4,994,803 (€2,812,348 plus £1,819,782) to fund my research since 2010 (this includes the funding awarded to my group only, no total grant value).
This is broken down into major project grants awarded as PI:
• 2025-27 HORIZON-MSCA-2024-PF-01 (EU, €260,348).
• 2024-28 UKRI CDT in AI for Healthcare (UK, 50% of £100,000).
• 2023-26 EPSRC Healthcare Technologies (UK, £625,936).
• 2022-27 Royal Society Wolfson Fellowship (UK, £281,280).
• 2021-24 Plancancer MIC (France, €276,493).
• 2020-23 ANR PRC grant (France, €320,000).
• 2019-22 CEFIPRA (India-France bilateral projects, €77,000).
• 2017-20 ANR Tremplin-ERC (France, €130,000).
• 2015-17 A*MIDEX Excellence Chair (France, €235,000).
• 2010-14 MRC Methodology Research Fellowship (UK, £507,936).
Project grants awarded as co-I:
• 2024-28 EPSRC CDT in Chemical Biology (UK, 20% of £250,000). PI: Ed Tate (Dept. Chemistry, Imperial).
• 2024-28 MRC-LMS Transdisciplinary Training Programme (UK, 50% of £170,000). PI: Jesus Gil (MRC LMS, Imperial).
• 2023-25 James West Scholarship for Ms Elizabeth Amelia (UK; £81,000).
• 2022 ARC – SIGN'IT 2022 (France, €110,000).
• 2022 ANR – PEPR Digital health (France, €500,000).
• 2016 Canceropôle PACA (France, €83,000).
And also awarded grants for postdoctoral fellowships (ARC, FRM), PhD scholarships (Chulabhorn Royal Academy, ANR Artificial Intelligence, IPC, HEC, CONACyT, PTDF, USTH) and other purposes (Pfizer, PHC Merlion, INSERM).
Publications and patents
As June 2023: 83 peer-reviewed papers since 2003, 79% as corresponding author. H-index restricted to papers where I am either first, last or corresponding author is 29.
Ballester, P.J. (2012) US Patent 8244483 “Shape Recognition Methods and Systems for Searching Molecular Databases”
Community service
Editorial roles: 2021-date Editorial Board Member of Briefings in Bioinformatics, 2019-date Associate Editor of npj Precision Oncology, among others. Peer-review: 500+ reviews of journal papers verified. Grant proposal evaluations for a range of funding agencies: Canada Research Chair, Medical Research Council (MRC, UK), DBT/Wellcome Trust India Alliance (India), Science Foundation (GACR, Czech Republic), National Research Foundation (NRF, South Africa), Ministry of Economy and Competitiveness (MINECO, Spain), Natural Sciences and Engineering Research Council (NSERC, Canada), Agency for Health Quality and Assessment of Catalonia (AQuAS, Spain), Engineering and Physical Sciences Research Council (EPSRC, UK), National Foundation for Science & Technology (FCT, Portugal), Health Research Board (HRB, Ireland), National Research Fund (FNR, Luxembourg), Israel Science Foundation (ISF), The Swiss National Science Foundation, Netherlands Organisation for Scientific Research, National Agency for Research (ANR, France), National Agency for Research (ANEP, Spain), Biotechnology and Biological Sciences Research Council (BBSRC, UK) and National Scientific Research Council (Romania). Times Higher Education’s World University Rankings: Invited to act as a reputation evaluator every year since 2018.
PhD students at Imperial (UK): In Bioengineering (2025: Chayanit Piyawajanusorn).
Postgraduate research interns at Imperial (UK): 2025: Hao Chen.
MRes students at Imperial (UK): In Bioengineering (2025: Victoria Medina; 2024: Cosmin Ichim, Sara Marsden, Jiatao Liu, Yuanjie Zou, Seshagiri Sakthimani), in Cancer Informatics (2025: Abhavya Raja), in Drug Discovery and Development (2025: Muhan Zhang), in Cancer Technology (2024: Tiantian He).
MSc students at Imperial (UK): In Bioengineering (2024: Alexis Dougha, Yue Cai; 2023: Lucas He, Alec Reygrobellet), in Digital Chemistry (2025: Naomi Ratnasingam; 2024: Dhyaksa Ariadi, Hannah Okesade; 2023: Amrit Jandu, Nicola Chesterman), in Bioinformatics (2023: Rasmus Hildebrandt).
Final-year undergraduate students at Imperial (UK): Betty Li (2025, MEng in BME), Charlotte Probstel (2025, MEng in BME), Mahad Parwaiz (2025, MEng in BME), Theo Sze (2025, MEng in MBE), Jonathan Talbot-Martin (2023, primarily supervised by Dr Nicky Whiffin, University of Oxford), Aparna Loecher (2022, primarily supervised by Dr Nuria Oliva-Jorge, Imperial College).
Postdocs at CRCM (France): Jitendra Kuldeep (2021, 18 months), Viet-Khoa Tran-Nguyen (2021, 2 years), Ghita Ghislat (2022, 9 months), Junaid Muhammad (2021, 18 months), Saw Simeon (2019, 2 years), Pavel Sidorov (2017, 2 years), Stefan Naulaerts (2017, 1.5 years), Cuong Dang (2015, 2 years), Antonio Peon (2015, 2 years).
PhD students at CRCM (France): Hongjian Li (visiting from CUHK), Linh Nguyen, Adeolu Ogunleye, Sachin Vishwakarma, Saiveth Hernández.
Master students at CRCM (France): Louai Kassa-Baghdouche, Dina Ouahbi, Pablo Gomez, Louison Fresnais, Amad Diouf, Michal Zulcinski, Nicola Jaume.
Master students at EMBL-EBI (UK): Raquel Romero, Saumya Kumar.
Examples of current alumni positions: lecturers/assistant professors (Linh Nguyen and Cuong Dang at Vietnam National University, Pavel Sidorov at Hokkaido University, Viet-Khoa Tran-Nguyen at Paris Cite University), pharmaceutical companies (Sachin Vishwakarma at Evotec), senior Horizon Europe Marie Curie fellowships (Ghita Ghislat at Imperial College London).
Collaborators
• Taufiq Rahman (Dept. of Pharmacology, University of Cambridge, UK): co-leading EPSRC project.
• Jesus Gil (MRC Laboratory of Medical Sciences, UK): co-supervising PhD student Seihee Jeong.
• Ed Tate, James Bull (Dept. of Chemistry, Imperial College London, UK), Oliver Schadt and Ingo Kober (Merck KGaA, Germany): co-supervising PhD student Daisy Williams.
• Nicholas Croucher (School of Public Health, Imperial College London, UK): co-supervising PhD student Joshua Fitch.
• Delphine Fradin (University of Nantes, France): collaborating with postdoc Dr Saiveth Hernandez.
• Richard Marchese-Robinson (Syngenta, UK): co-supervising PhD student Hannah Okesade.
Teaching
• 2024-26 module creator, leader and lecturer of Artificial intelligence for drug discovery (BIOE70150).
• 2023-26 module and element leader of Bioengineering Group Project (BIOE60005).
• 2023-24 module leader of Mathematics 1 (BIOE40004) and Maths and Engineering 1 (BIOE40005).
• 2022-26 module lecturer of Mathematics 1 (BIOE40004) and Maths and Engineering 1 (BIOE40005).
• 2022-24 guest lecturer of Nanotechnologies for cancer diagnosis and cancer therapy (BIOE97173).
• 2016 Introductory talk to first-year master students engineering students from École Centrale de Marseille on how computational modelling can be applied to cancer research.
• 2010-2014 Science Society member, Wolfson College Cambridge: Inviting public lecture speakers, organising their lectures and entertaining them during their visit.
• 2013-14 guest lecturer at UCL's MSc in Drug Design, on computational methods for drug design.
• 2013 guest lecturer at Cambridge's Computational Biology Institute on ML approaches to predicting protein-ligand binding.
• 2011 Organised a 1-day visit to GSK for EMBL-EBI and UCL postgraduates: lectures, networking and career opportunities in industry.
News
Join us
We are always interested in hearing from prospective group members in any area directly related to our research interests (computational chemistry, bioinformatics, machine learning, etc.). Please email p.ballester@imperial.ac.uk with your CV, grades from degrees, publications and the funding opportunity you would like to apply for. Note that I have to be contacted well ahead of the deadline, as we have to apply for departmental support as well. Some funding opportunities per career stage are provided below:
Postdocs
• Royal Commission 1851
• Leverhulme Trust
• Marie Sklodowska-Curie Individual Fellowship
• British Council Women in STEM Fellowship (South Asia)
• HFSP Postdoctoral Fellowships
PhD student
• President's PhD Scholarship Scheme
• China Scholarship Council (China)
• Imperial Marshall Scholarship (USA)
• All Scholarship Schemes (including a scholarship search tool)
• Bioengineering Department Studentships
• Conicyt-Imperial Scholarship (Chile)
• High income Commonwealth PhD Scholarships (eligible countries within)
• Commonwealth PhD Scholarships (eligible countries within)
• Commonwealth Split-site Scholarship (eligible countries within)
Information about the PhD programme in the Department of Bioengineering can be found here. For general information on the tuition fees and cost of living in London, please read this link. For other sources of PhD funding you can also have a look here, here (BioEngineering funding) and here (fees and funding). Some external scholarships can also be found here. Before contacting me, please check that you fulfill the pre-requisites for application and entry set by Imperial College.
Undergraduate or master students
You only have to email me your CV, grades from degrees, and course details.
Contact
Email: p.ballester@imperial.ac.uk
Address: Sir Michael Uren Hub, Imperial College White City campus, 86 Wood Lane, London - W12 0BZ, UK
How to get here: https://www.imperial.ac.uk/visit/campuses/white-city/
About this campus: https://www.imperial.ac.uk/white-city-campus/